| X | ID_OSU | Gender | Weight | Beers | BAC |
|---|---|---|---|---|---|
| 1 | 1 | female | 132 | 5 | 0.100 |
| 2 | 2 | female | 128 | 2 | 0.030 |
| 3 | 3 | female | 110 | 9 | 0.190 |
| 4 | 4 | male | 192 | 8 | 0.120 |
| 5 | 5 | male | 172 | 3 | 0.040 |
| 6 | 6 | female | 250 | 7 | 0.095 |
| 7 | 7 | female | 125 | 3 | 0.070 |
| 8 | 8 | male | 175 | 5 | 0.060 |
| 9 | 9 | female | 175 | 3 | 0.020 |
| 10 | 10 | male | 275 | 5 | 0.050 |
| 11 | 11 | female | 130 | 4 | 0.070 |
| 12 | 12 | male | 168 | 6 | 0.100 |
| 13 | 13 | female | 128 | 5 | 0.085 |
| 14 | 14 | male | 246 | 7 | 0.090 |
| 15 | 15 | male | 164 | 1 | 0.010 |
| 16 | 16 | male | 175 | 4 | 0.050 |
Day 40
Math 216: Statistical Thinking
Blood Alcohol Content (BAC)
Scatter Plot of BAC vs Beers
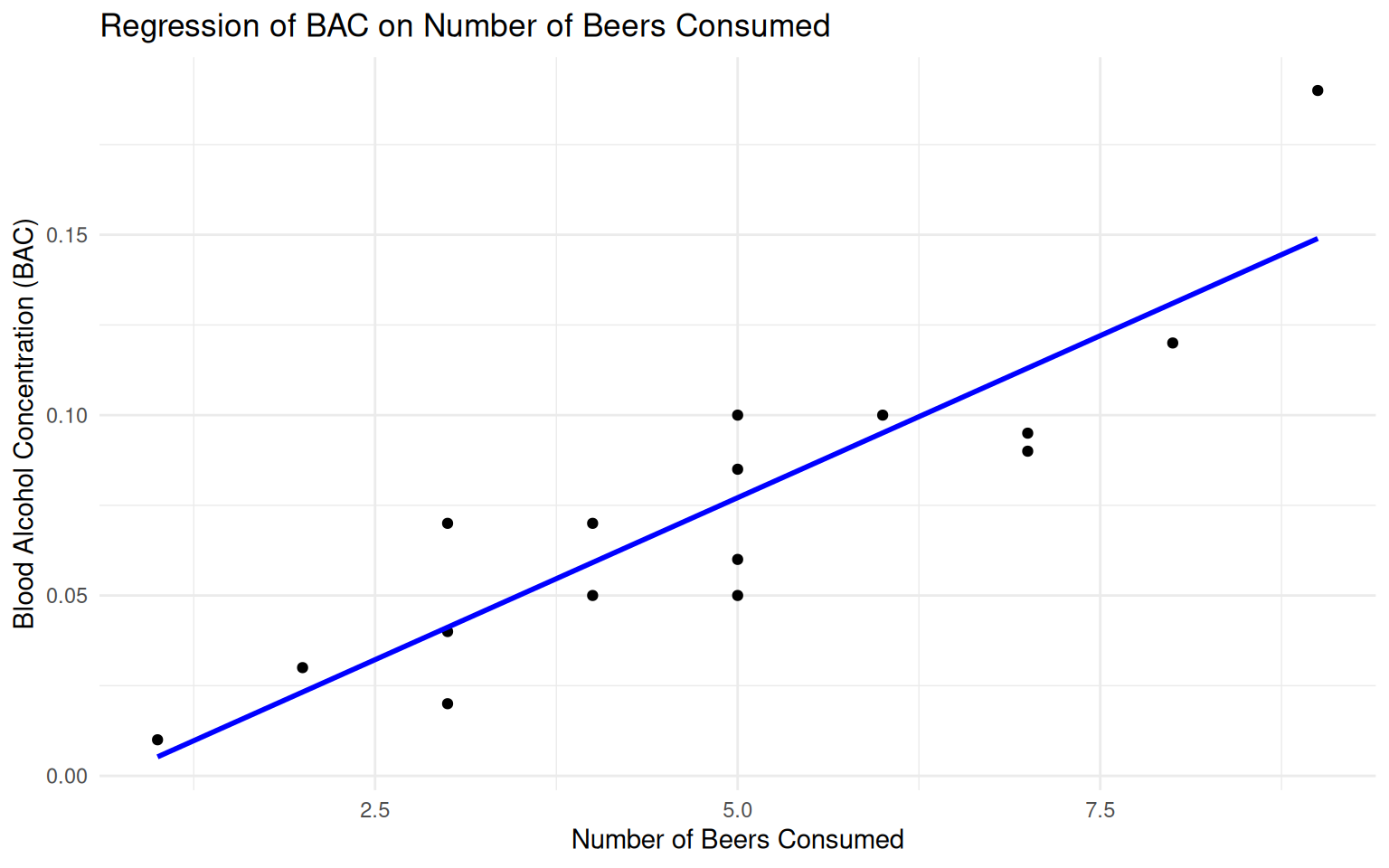
Simple Linear Regression of BAC (y) on Beers (x)
Key Question: How does beer consumption affect blood alcohol content?
\[ \begin{align*} \widehat{BAC} &=-0.0127+0.0180(\text{Beers})\\ \hat{\sigma} &= 0.02044 \end{align*} \]
Real-World Insight: Each additional beer increases BAC by about 0.018 units on average!
Call:
lm(formula = BAC ~ Beers, data = bac)
Residuals:
Min 1Q Median 3Q Max
-0.027118 -0.017350 0.001773 0.008623 0.041027
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.012701 0.012638 -1.005 0.332
Beers 0.017964 0.002402 7.480 2.97e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.02044 on 14 degrees of freedom
Multiple R-squared: 0.7998, Adjusted R-squared: 0.7855
F-statistic: 55.94 on 1 and 14 DF, p-value: 2.969e-06Confidence Interval
Key Question: How precise are our slope and intercept estimates?
\[ C\% \text{ confidence interval for } \beta_i \text{ is } \hat{\beta}_i \pm t^* \operatorname{SE}(\hat{\beta}_i) \]
- Get CIs for slope/intercept with
confintcommand or compute usingqt(.975, df= )to get t* for 95% CI
Real-World Insight: This tells us the range of plausible values for the true effect of beers on BAC!
2.5 % 97.5 %
(Intercept) -0.03980535 0.01440414
Beers 0.01281262 0.02311490Inference for slope (effect of Beers on BAC)
Key Question: Is the relationship between beers and BAC statistically significant?
\[ \begin{align*} \mathrm{H}_0: &\ \beta_i = 0 & \text{(no effect for predictor i)} \\ \mathrm{H}_A: &\ \beta_i \neq 0 & \text{(predictor i has an effect on y)} \end{align*} \]
Real-World Insight: This test tells us if we’re seeing a real effect or just random variation!
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -0.0127006 | 0.0126375 | -1.004993 | 0.3319551 |
| Beers | 0.0179638 | 0.0024017 | 7.479592 | 0.0000030 |
Multiple Regression Framework
Key Question: How do we extend simple regression to handle multiple predictors?
Multiple Regression Model: \[ Y = \beta_0 + \beta_1 X_1 + \beta_2 X_2 + \cdots + \beta_p X_p + \varepsilon \]
Interpretation: Each coefficient \(\beta_i\) represents the change in \(Y\) for a one-unit change in \(X_i\), holding all other predictors constant
Assumptions: Same as simple regression but extended to multiple dimensions
Real-World Insight: Multiple regression lets us control for confounding variables and understand complex relationships!
Multiple Regression Decision Framework
%%{init: {"theme": "base", "themeVariables": {"fontSize": "16px", "fontFamily": "Arial", "lineColor": "#333"}}}%%
flowchart TD
%% --- Styling Definitions ---
classDef start fill:#FFFACD,stroke:#FF8C00,stroke-width:3px,color:#000
classDef decision fill:#E6F3FF,stroke:#1E88E5,stroke-width:3px,color:#000
classDef action fill:#E8F5E9,stroke:#43A047,stroke-width:3px,color:#000
classDef endStyle fill:#FFEBEE,stroke:#E53935,stroke-width:3px,color:#000
%% --- Diamond Pattern Layout ---
A["Research Question"]:::start
B["Identify Predictors<br/>Select X₁, X₂, ..., Xₖ"]:::action
C["Fit Multiple Regression<br/>Y = β₀ + β₁X₁ + β₂X₂ + ...+ βₖXₖ + ε<br/>where ε ~ N(0, σ²)"]:::action
D["Check Assumptions"]:::decision
E["t-Test Coefficients<br/>t = β̂ⱼ/SE(β̂ⱼ)<br/>H₀: βⱼ = 0"]:::action
F["F-Test Overall Model<br/>F = (SSR/k)/(SSE/(n-k-1))<br/>R² = SSR/SST"]:::action
G["Interpret Results<br/>95% CI: β̂ⱼ ± t*·SE(β̂ⱼ)"]:::action
%% --- Diamond Pattern Connections ---
A --> B
B --> C
C --> D
D --> E
D --> F
E --> G
F --> G
%% --- Visual Polish ---
linkStyle default stroke:#333,stroke-width:3px;
linkStyle 3 stroke:#333,stroke-width:3px;
linkStyle 4 stroke:#333,stroke-width:3px;
linkStyle 5 stroke:#333,stroke-width:3px;
linkStyle 6 stroke:#333,stroke-width:3px;
Scatterplot Matrix for BAC Example
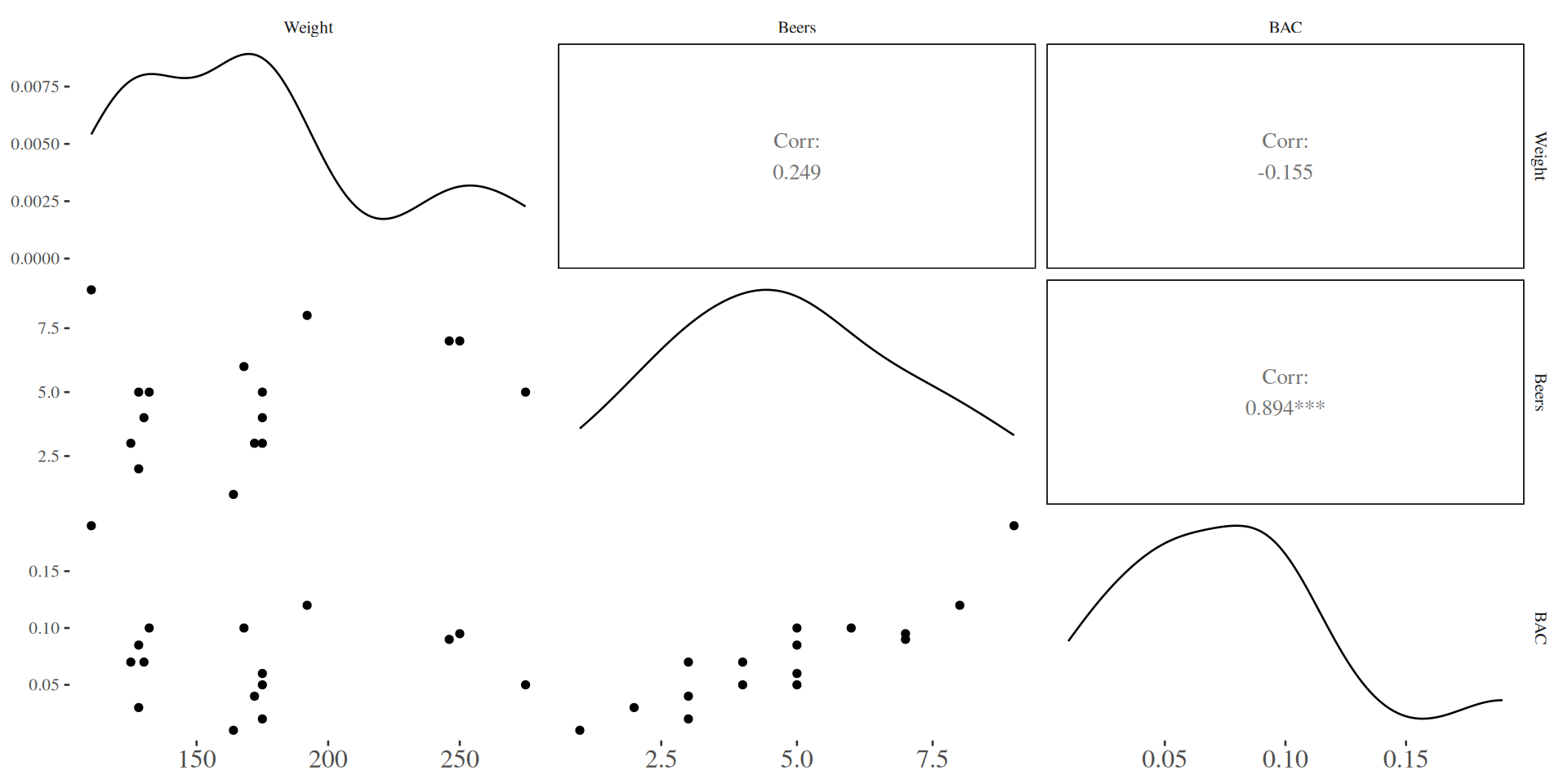
Regression of BAC on Beers and Weight
The fitted model for BAC is:
Call:
lm(formula = BAC ~ Beers + Weight, data = bac)
Residuals:
Min 1Q Median 3Q Max
-0.0162968 -0.0067796 0.0003985 0.0085287 0.0155621
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.986e-02 1.043e-02 3.821 0.00212 **
Beers 1.998e-02 1.263e-03 15.817 7.16e-10 ***
Weight -3.628e-04 5.668e-05 -6.401 2.34e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.01041 on 13 degrees of freedom
Multiple R-squared: 0.9518, Adjusted R-squared: 0.9444
F-statistic: 128.3 on 2 and 13 DF, p-value: 2.756e-09\[ \widehat{BAC} = 0.0399 + 0.0200 (\text{Beers}) - 0.00036 (\text{Weight}). \]
Regression of BAC on Beers and Weight
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 0.0398634 | 0.0104333 | 3.820787 | 0.0021219 |
| Beers | 0.0199757 | 0.0012629 | 15.817343 | 0.0000000 |
| Weight | -0.0003628 | 0.0000567 | -6.401230 | 0.0000234 |
| 2.5 % | 97.5 % | |
|---|---|---|
| (Intercept) | 0.0173236 | 0.0624031 |
| Beers | 0.0172474 | 0.0227040 |
| Weight | -0.0004853 | -0.0002404 |
Regression of BAC on Beers, Weight, and Gender
Key Question: How do all three factors together predict BAC?
\[ \widehat{BAC} = 0.039 + 0.020 (\text{Beers}) - 0.00034 (\text{Weight}) - 0.0032 \text{ (Male)} \]
Real-World Insight: This model shows that gender matters too - males have slightly lower BAC than females with the same beer consumption and weight!
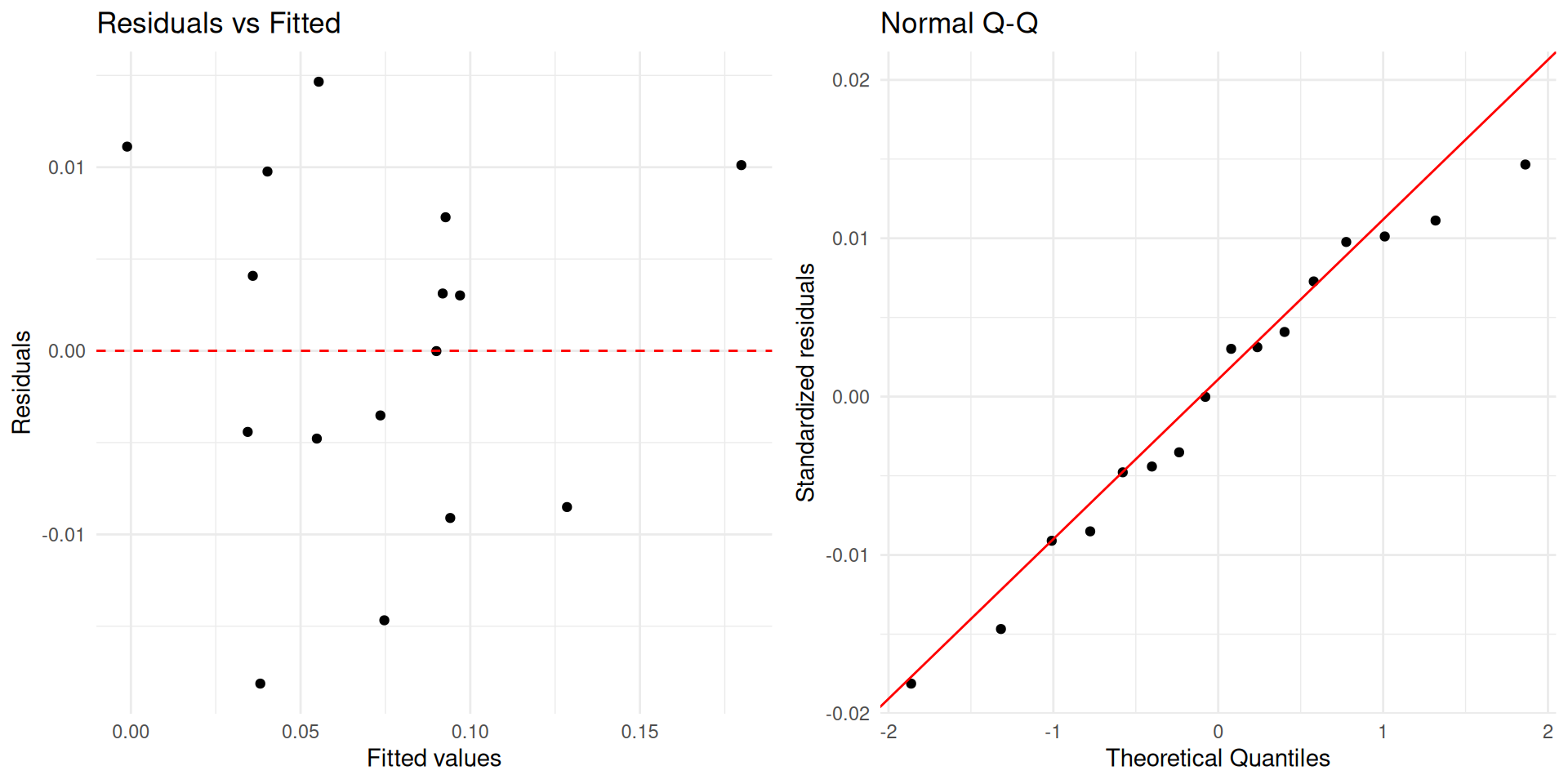
Both number of beers and weight are statistically significant predictors of BAC (p-value < 0.0001). Holding weight constant, we are 95% confident that the true effect of drinking one more beer is a 0.017 to 0.023 unit increase in mean BAC.
Scatterplot Matrix for BAC Example
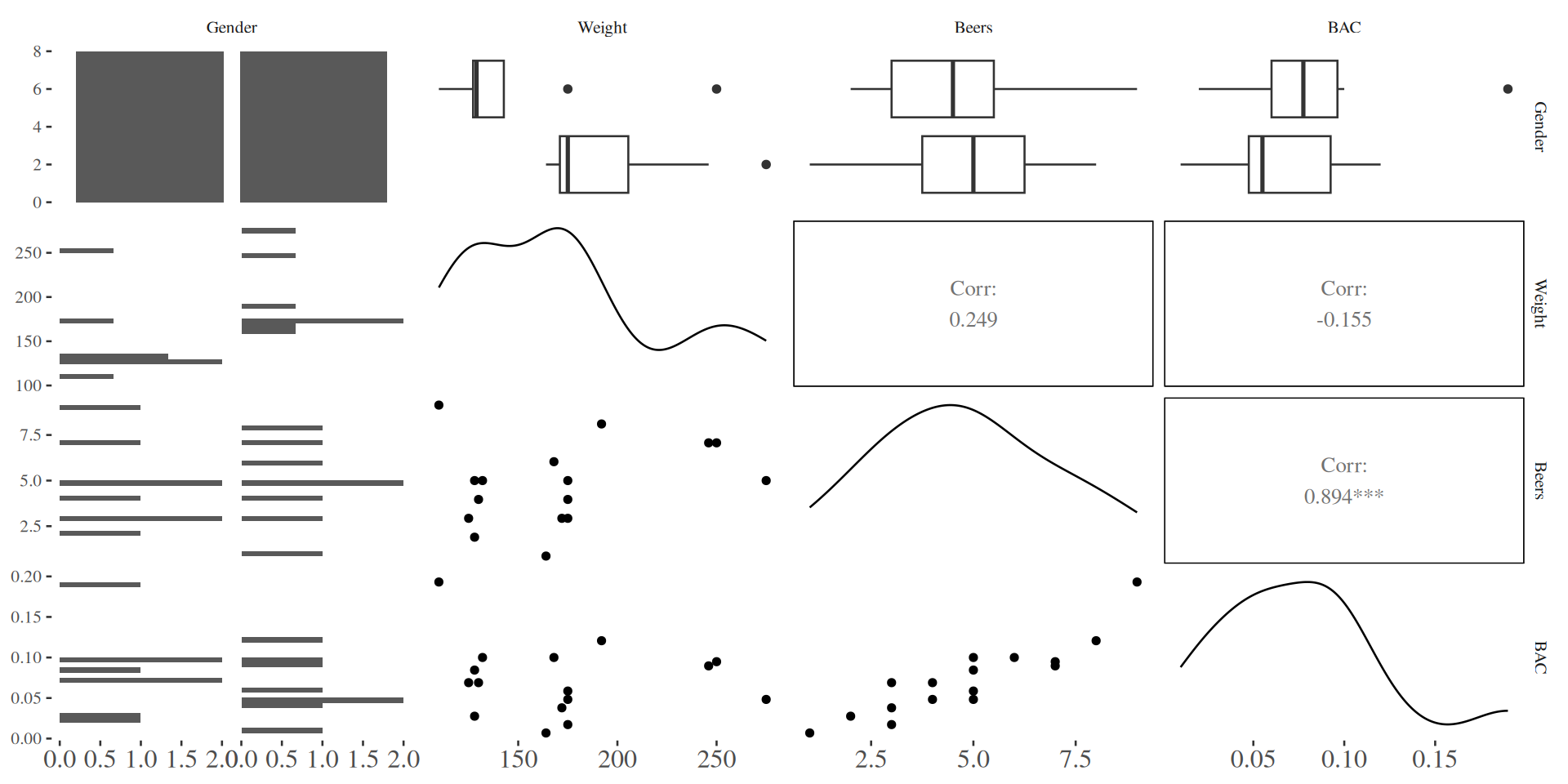
Regression for BAC on Beers, Weight, and Gender
Call:
lm(formula = BAC ~ Beers + Weight + Gender, data = bac)
Residuals:
Min 1Q Median 3Q Max
-0.018125 -0.005713 0.001501 0.007896 0.014655
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.871e-02 1.097e-02 3.528 0.004164 **
Beers 1.990e-02 1.309e-03 15.196 3.35e-09 ***
Weight -3.444e-04 6.842e-05 -5.034 0.000292 ***
Gendermale -3.240e-03 6.286e-03 -0.515 0.615584
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.01072 on 12 degrees of freedom
Multiple R-squared: 0.9528, Adjusted R-squared: 0.941
F-statistic: 80.81 on 3 and 12 DF, p-value: 3.162e-08Regression Details for BAC on Beers, Weight, and Gender
“Male” is an indicator variable that equals 1 when predicting male Blood Alcohol Content (BAC) and 0 for female.
- Barb’s Prediction
- Context: Barb drank 4 beers, weighs 160 lbs, and is female.
- Equation: \(\widehat{BAC} = 0.039 + 0.020(4) - 0.00034(160) - 0.0032(0) = 0.0646\)
- John’s Prediction
- Context: John drank 4 beers, weighs 160 lbs, and is male.
- Equation: \(\widehat{BAC} = 0.039 + 0.020(4) - 0.00034(160) - 0.0032(1) = 0.0614\)
Real-World Insight: These examples show how the model accounts for individual differences - same beer consumption but different BAC predictions!
Calculation Verification
Key Question: How do we verify our multiple regression calculations in R?
- Manual Prediction:
# Manual calculation verification
barb_pred_manual <- 0.039 + 0.020*4 - 0.00034*160 - 0.0032*0
john_pred_manual <- 0.039 + 0.020*4 - 0.00034*160 - 0.0032*1
# R prediction - ensure factor levels match
barb_data <- data.frame(Beers = 4, Weight = 160, Gender = factor("F", levels = levels(bac$Gender)))
john_data <- data.frame(Beers = 4, Weight = 160, Gender = factor("M", levels = levels(bac$Gender)))Barb's BAC: Manual: 0.0646 John's BAC: Manual: 0.0614 Model Disgnostics